AML has three projects:
1.
Trajectory and Architecture of Tumor Intrinsic Drug Resistance in AML.
2.
Trajectory and Architecture of Microenvironment-Mediated Resistance in AML.
3.
Translating Improved Pairing and Timing of Drug Combination Strategies.
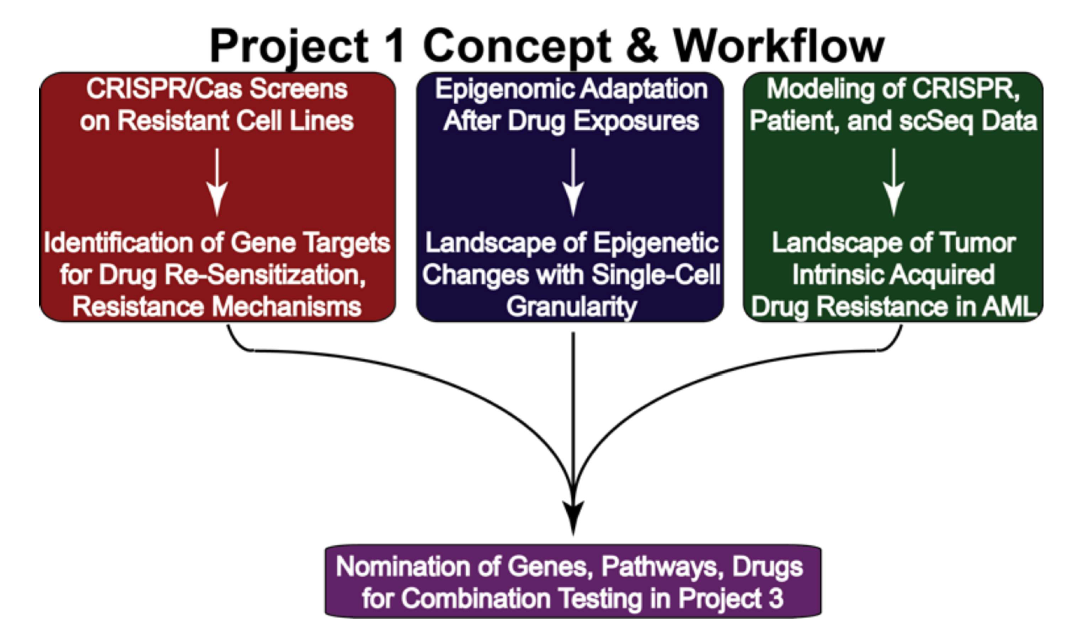
The long-term goal of this project is to optimize and bring the most effective drug combinations into clinical use for patients with acute myeloid leukemia (AML). The immediate objective is to understand the tumor-intrinsic mechanisms behind acquired drug resistance. To achieve these goals, three specific aims have been outlined:
1.
Next-generation genome-wide interrogation of key acquired resistance scenarios: A panel of AML models with acquired drug resistance has been created through long-term drug exposure, sometimes supplemented with extrinsic cytokines. These drug-resistant cells will undergo genome-wide CRISPR screening with various drugs or drug combinations to identify key resistance mechanisms.
2.
Epigenomic evolution of acquired resistance: Protocols will be used to expand myeloid progenitor cells from primary AML patient samples to study how epigenetic changes contribute to resistance. Single-cell sequencing will be employed to profile shifts in the epigenetic landscape in response to the same drugs and combinations used in Aim 1.
3.
Atlas of intrinsic drug resistance in AML: Leveraging expertise in data integration and modeling, the existing functional genomic dataset will be combined with new data from Aims 1 and 2 to create an Atlas of tumor-intrinsic mechanisms of acquired drug resistance in AML.
These innovative analyses are expected to significantly enhance the understanding of acquired drug resistance in AML, ultimately leading to the successful clinical application of more effective drug combination strategies.
(For additional information, please visit NIH RePORTER)
The team's central hypothesis is that AML tumors adapt to therapeutic pressure through a multi-step process, initially involving interaction with immune and stromal cells, eventually leading to a resistant state characterized by clonal evolution. The project's long-term goal is to optimize and introduce effective drug combinations into clinical practice for AML patients. The immediate goal is to understand the tumor-intrinsic mechanisms of acquired drug resistance, addressed through three specific aims:
1.
Next-generation genome-wide interrogation of key acquired resistance scenarios: The team created AML models with acquired drug resistance through long-term drug exposure and will perform CRISPR screens on these models to identify resistance mechanisms.
3.
Epigenomic evolution of acquired resistance: They will expand myeloid progenitor cells from AML patient samples to study how epigenetic changes contribute to resistance, using single-cell sequencing to profile shifts in response to drugs.
3.
Atlas of intrinsic drug resistance in AML: The team will integrate existing and new data to create an Atlas of tumor-intrinsic mechanisms of resistance, using advanced data modeling approaches.
These innovative efforts aim to deepen the understanding of drug resistance in AML and pave the way for more effective clinical treatments.
(For additional information, please visit NIH RePORTER)
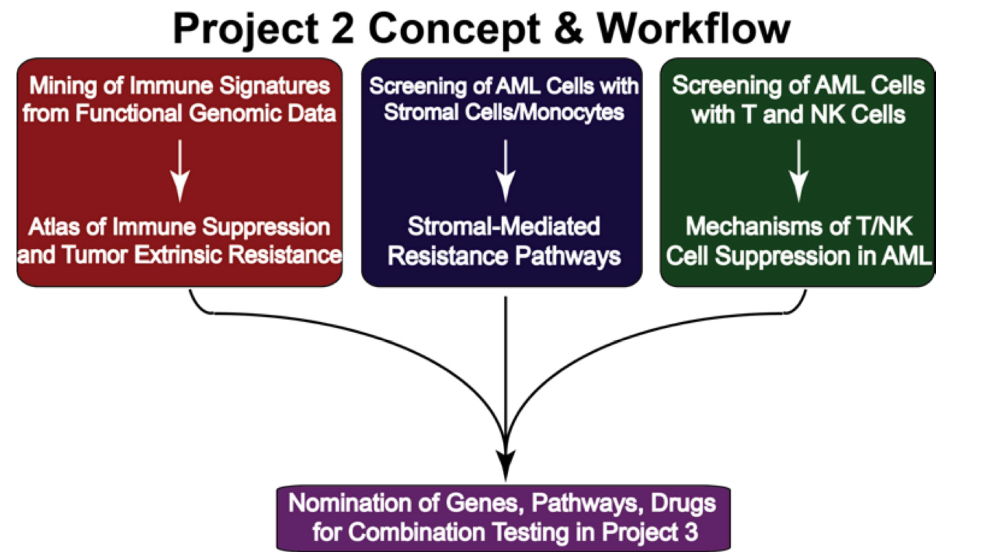
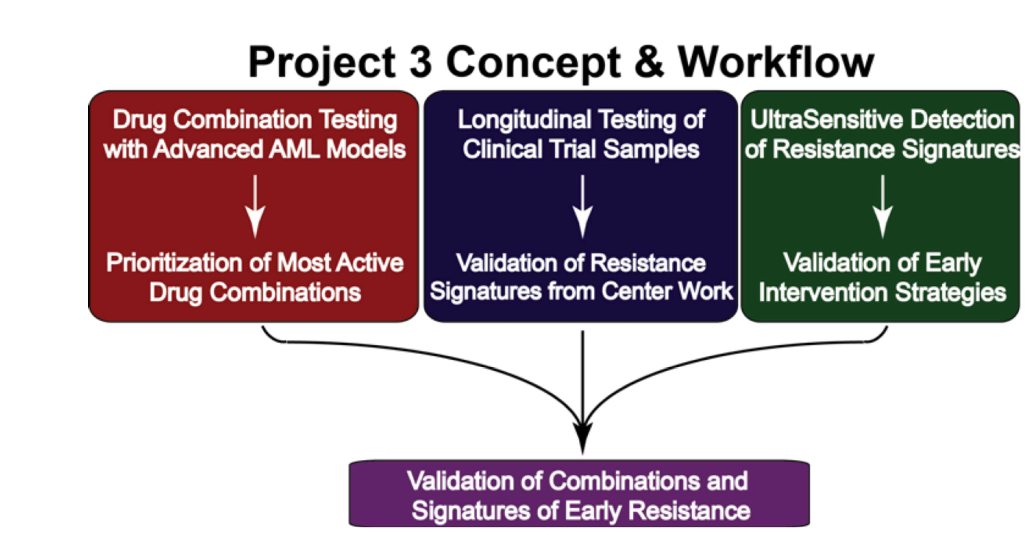
The goal of this project is to develop and implement new drug combinations to prevent relapse and improve outcomes for acute myeloid leukemia (AML) patients by targeting mechanisms of acquired drug resistance. Previous research has shown that AML relapse is driven by tumor cells adapting with support from the bone marrow microenvironment, leading to drug resistance through a multi-stage process. Early resistance is triggered by external signals and eventually evolves into a more advanced, clonal resistance. This understanding opens opportunities for early intervention to prevent resistance.
The project's immediate objectives are to prioritize promising drug combinations and reliable resistance markers for clinical use. To achieve these goals, three specific aims are proposed:
1.
Evaluate signatures of resistance using primary AML samples in ex vivo assays: The team will use their expertise in testing primary AML patient samples against drug combinations using high-throughput screening, imaging, flow-based readouts, and a 3D bone marrow model (Humarrow) for long-term drug testing.
2.
Longitudinal evaluation of samples from patients receiving rational therapeutic regimens: Detailed analysis will be performed on samples from patients in clinical trials to identify resistance signatures at early therapy stages.
3.
Employ sensitive detection techniques to detect low levels of resistance in primary samples: Advanced single-cell and cell enrichment techniques will be used to detect early resistance markers in newly diagnosed and early-stage AML patients.
This work aims to refine clinical trials and identify new treatments to prevent disease relapse and improve patient outcomes by addressing resistance at the earliest stages.
(For additional information, please visit NIH RePORTER)